COMPUTER VISION PROJECTS
 |
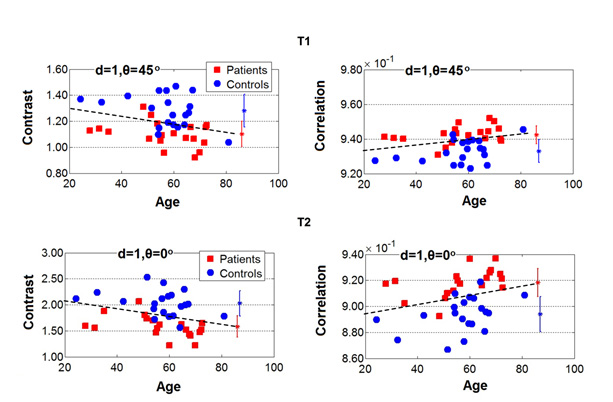
Corticospinal tract degeneration in ALS unmasked in T1-weighted images using texture analysis The purpose of this study was to investigate whether textures computed from T1-weighted (T1W) images of the corticospinal tract (CST) in amyotrophic lateral sclerosis (ALS) are associated with degenerative changes evaluated by diffusion tensor imaging (DTI). Nineteen patients with ALS and 14 controls were prospectively recruited and underwent T1W and diffusionweighted magnetic resonance imaging. Three-dimensional texture maps were computed from T1W images and correlated with the DTI metrics within the CST. Significantly correlated textures were selected and compared within the CST for group differences between patients and controls using voxel-wise analysis. Textures were correlated with the patients' clinical upper motor neuron (UMN) signs and their diagnostic accuracy was evaluated. Voxel-wise analysis of textures and their diagnostic performance were then assessed in an independent cohort with 26 patients and 13 controls. Results showed that textures autocorrelation, energy, and inverse difference normalized significantly correlated with DTI metrics (p < .05) and these textures were selected for further analyses. The textures demonstrated significant voxel-wise differences between patients and controls in the centrum semiovale and the posterior limb of the internal capsule bilaterally (p < .05). Autocorrelation and energy significantly correlated with UMN burden in patients (p < .05) and classified patients and controls with 97% accuracy (100% sensitivity, 92.9% specificity). In the independent cohort, the selected textures demonstrated similar regional differences between patients and controls and classified participants with 94.9% accuracy. These results provide evidence that T1-based textures are associated with degenerative changes in the CST. Reference A. Ishaque, D. Mah, P. Seres, C. Luk, W. Johnston, S. Chenji, C. Beaulieu, Y.H. Yang, and S. Kalra, Human Brain Mapping, 2018. |
|
 |
Evaluating the cerebral correlates of survival in amyotrophic lateral sclerosis Objective: To evaluate cerebral degenerative changes in ALS and their correlates with survival using 3D texture analysis. Methods: A total of 157 participants were included in this analysis from four neuroimaging studies. Voxel-wise texture analysis on T1-weighted brain magnetic resonance images (MRIs) was conducted between patients and controls. Patients were divided into long- and short-survivors using the median survival of the cohort. Neuroanatomical differences between the two survival groups were also investigated. Results: Whole-brain analysis revealed significant changes in image texture (FDR P < 0.05) bilaterally in the motor cortex, corticospinal tract (CST), insula, basal ganglia, hippocampus, and frontal regions including subcortical white matter. The texture of the CST correlated (P < 0.05) with finger- and foot-tapping rate, measures of upper motor neuron function. Patients with a survival below the media of 19.5 months demonstrated texture change (FDR P < 0.05) in the motor cortex, CST, basal ganglia, and the hippocampus, a distribution which corresponds to stage 4 of the distribution TDP-43 pathology in ALS. Patients with longer survival exhibited texture changes restricted to motor regions, including the motor cortex and the CST. Interpretation: Widespread gray and white matter pathology is evident in ALS, as revealed by texture analysis of conventional T1-weighted MRI. Length of survival in patients with ALS is associated with the spatial extent of cerebral degeneration. Reference A. Ishaque, D. Mah, P. Seres, C. Luk, D. Eurich, W. Johnston, Y.H. Yang, and S. Kalra, Ann. Clinical and Trans. Neurology, 2018. |
|
 |
Texture Analysis to Detect Cerebral Degeneration in Amyotrophic Lateral Sclerosis Background: Evidence of cerebral degeneration is not apparent on routine brain MRI in amyotrophic lateral sclerosis Reference A. Ishaque, R. Maani, J. Satkunam, P. Seres, D. Mah, A.H. Wilman, S. Naik, Y.H. Yang, and S. Kalra, Can. J. Neuro. Sci., Vol. 45, 2018, pp. 533-539. |
|
Statistical map of local texture features (f7 and f8) corrected by FDR at p<0.01 in the AD database. |
Voxel-Based Texture Analysis of the Brain This project presents a novel voxel-based method for texture analysis of brain images. Texture analysis is a powerful quantitative approach for analyzing voxel intensities and their interrelationships, but has been thus far limited to analyzing regions of interest. The proposed method provides a 3D statistical map comparing texture features on a voxel-by-voxel basis. The validity of the method was examined on artificially generated effects as well as on MRI data in Alzheimer's Disease (AD). The artificially generated effects included hyperintense and hypointense signals added to T1-weighted brain MRIs from 30 healthy control subjects. The proposed method detected artificial effects with high accuracy and revealed statistically significant differences between the AD and control groups. This project extends the usage of texture analysis byond the current region of interest analysis to voxel-by-voxel 3D statistical mapping and provides a hypothesis-free analysis tool to study cerebral pathology in neurological diseases. Reference R. Maani, Y. H. Yang, and S. Kalra, "Voxel-Based Texture Analysis of the Brain," PLOS One, March, 2015. |
|
 |
Robust Volumetric Texture Classification of MRI of the Brain using Local Frequency Descriptor This project presents a new method for robust volumetric texture classification. It also proposes 2D and 3D gradient calculation methods designed to be robust to imaging effects and artifacts. Using the proposed 2D method, the gradient information is extracted on the XYZ orthogonal planes at each voxel and used to form a local coordinate system. The local coordinate system and the local 3D gradient computed by the proposed 3D gradient calculator are then used to define volumetric texture features. It is shown that the presented gradient calculation methods can be efficiently implemented by convolving with 2D and 3D kernels. The experimental results demonstrate that the proposed gradient operators and the texture features are robust to imaging effects and artifacts, such as blurriness and noise in 2D and 3D images. The proposed method is compared with 3 state-of-the-art volumetric texture classification methods, namely, the 3D GLCM, 3D LBP, and second orientation pyramid on MRI data of the brain. The experimental results show the superiority of the proposed method in accuracy, robustness and speed. Reference R. Maani, S. Kalra, and Y.H. Yang, "Robust Volumetric Texture Classification of Magnetic Resonance Images of the Brain Using Local Frequency Descriptor," IEEE Trans. on Image Processing, Vol. 23, No. 10, 2014, pp. 4625-4636. |
|
 |
Texture Analysis of the Brain in ALS Amyotrophic Lateral Sclerosis (ALS) is a fatal neurodegenerative disease characterized by progressive muscular paralysis. The diagnosis of ALS is based on the clinical assessment of both upper motor neuron (UMN) and lower motor neuron (LMN) degeneration. While the detection of LMN dysfunction is aided by electrophysiologic techniques, an objective and accurate method to detect and quantify UMN degeneration is greatly needed. Reference R. Maani, Y.H. Yang, and S. Kalra, "Texture Analysis of the Brain in Amyotrophic Lateral Sclerosis," 19th Annual Meeting of the Organization for Human Brain Mapping," June 16-20, 2013, Seattle, WA. Acknowledgments This project has been funded by NSERC, ALS Canada, and the ALS Association. |
|
A reconstructed fetus |
3D Ultrasound In this project, we develop techniques for the analysis and visualization of ultrasound images. For analysis, we employ texture to perform classification and segmentation. For visualization, we employ optical flow to reconstruct in 3D the surface profile of a fetus. References R. Muzzolini, Y.H. Yang and R. Pierson, "Classifier design with incomplete knowledge," Pattern Recognition, Vol. 31, No. 4, 1998, pp. 345-369. (won the Honourable Mention Award for Best Paper by the Pattern Recognition Society) N. Weng, Y.H. Yang, and R. Pierson, "Three-dimensional surface reconstruction using optical flow for medical imaging," IEEE Trans. on Medical Imaging, Vol. 16, No. 5, 1997, pp. 630-641. R. Muzzolini, Y.H. Yang and R. Pierson, "Texture characterization using robust statistics," Pattern Recognition, Vol. 27, No. 1, 1994, pp. 119-134. R. Muzzolin, Y.H. Yang and R. Pierson, "Multiresolution texture segmentation with application to diagnostic ultrasound images," IEEE Trans. on Medical Imaging, Vol. 12, No. 1, 1993, pp. 108-123.
|
|
|
|