greedy algorithms
mst
definitions
graph, path, connected, component, cycle, forest, tree, spanning, weighted graph, mst
tree properties
removing cycle edge does not change components
why?
tree with at least 2 nodes has at least 2 nodes with degree 1
ends of longest path
tree with n nodes has n-1 edges
induction, ends of longest path
connected, n node, n-1 edges implies tree
must be node with degree 1
tree if and only if, between each node pair, unique path
ends of longest path
T spanning tree of G, e not in T, then
T+e has cycle C
for every c in C, T+e-c spanning tree
if T mst, then w(e) ≥ w(c)
X edge subset of mst T, S node subset of V such that no x crosses cut (S,V-X), e lightest edge across cut, then X+e edge subset of mst T’
Prim's mst
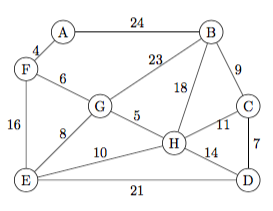
A B C D E F G H
tree *
E update 21 E * 16 E 8 E 10 E
GE 8 * *
G update 23 G 21 E * 6 G * 5 G
HG 5 * * *
H update 18 H 11 H 14 H * 6 G * *
FG 6 * * * *
F update 4 F 18 H 11 H 14 H * * * *
AF 4 * * * * *
A update * 18 H 11 H 14 H * * * *
CH 11 * * * * * *
C update * 9 C * 7 C * * * *
DC 7 * * * * * * *
D update * 9 C * * * * * *
BC 9 * * * * * * * *
def prim(G,source):
cost, parent, fringe = {}, {}, []
infin = weighted.infinity(G)
for v in G:
cost[v], parent[v] = infin, -1
fringe.append(v)
cost[source],parent[source] = 0, source
sum = 0
print source, 'start'
while len(fringe)>0:
u = fringe.pop(weighted.indexOfMin(fringe,cost))
sum += cost[u]
if u != source: print u,parent[u],cost[u]
if cost[u] == infin: doneEarly = True
for (v,costuv) in G[u]:
if (v in fringe) and (cost[v] > costuv):
cost[v] = costuv
parent[v] = u
print 'mst weight', sum
prim(weighted.G4,'A')
Kruskal's mst

AF 4 GH 5 FG 6 CD 7 EG 8 BC 9 reject EH CH 11 reject DH reject EF reject BH reject DE reject BG reject AB
what component are you in?
what component are you in?
merge components containing x,y
leads to a union-find data structure
simplest implementation
component: tree, with parent pointers
find(a): return root of tree containing a
union(a,b): parent[root(a)] = root(b)
simple union find data structure
def myfind(v,P): # simple find
while P[v] != v:
v = P[v]
return v
def myunion(x,y,P): # simple union
rootx, rooty = myfind(x,P), myfind(y,P)
P[rootx] = rooty
import weighted, UF
# convert from weighted adjacency list
# to tuple list: (v,w,wt_vw)
def createEdgeList(G):
L = []
for v in G:
for (w,wt) in G[v]:
if ord(v)<ord(w): L.append((v,w,wt))
return L
# extract tuple with min wt
def extractmin(L):
ndxMin, valMin = 0, L[0][2]
for j in range(1,len(L)):
t = L[j]
if t[2] < valMin:
ndxMin, valMin = j, t[2]
return L.pop(ndxMin)
def kruskalDemo(G):
L = createEdgeList(G)
parent = {}
for v in G: parent[v] = v
while len(L) > 0:
t = extractmin(L)
a, b = t[0], t[1]
ra, rb = UF.myfind(a,parent), UF.myfind(b,parent)
print a, ra, b, rb,
if ra != rb:
print 'add edge',a,b,t[2]
UF.myunion(ra,rb,parent)
else:
print 'reject ',a,b
kruskalDemo(weighted.G4)
a r b s r,s: roots of UF comp'ts with a,b
parent
A B C D E F G H
A A F F add A F 4 F B C D E F G H
G G H H add G H 5 F B C D E F H H
F F G H add F G 6 F B C D E H H H
C C D D add C D 7 F B D D E H H H
E E G H add E G 8 F B D D H H H H
B B C D add B C 9 F D D D H H H H
E H H H reject E H
C D H H add C H 11 F D D H H H H H
stop: n-1 edges
final UF structure: H
F G E D
A C B
Kruskal mst correctness
Claim: tree returned by Kruskal is mst.
Proof (sketch)
at each step, set of picked edges is acyclic, so a forest.
number of components of n-node m-edge acyclic forest is n-m (exercise: PBI)
so, when number picked edges < n-1, still ≥2 components. since input graph connected, must be an edge in input graph with ends different forest components. so, K's alg can pick some edge.
so K's alg picks n-1 edges, leaving acyclic graph, n nodes, 1 component, so spanning acyclic component, so spanning tree.
Claim: the tree returned is an MST.
Proof (sketch):
Let T be the tree returned by Kruskal's algorithm. Let T* be an MST. If T=T* we are done, so assume T ≠ T*.
Label the edges of T
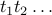 in the order they are selected by
Kruskal's algorithm, i.e. non-decreasing weight.
in the order they are selected by
Kruskal's algorithm, i.e. non-decreasing weight.Let k be the smallest index such that
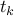 is not in T*.
is not in T*.Notice that T*
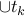 has a cycle C containing
has a cycle C containing 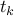 ,
and that removing any edge of C leaves a spanning tree
,
and that removing any edge of C leaves a spanning tree why? because the resulting graph is connected, and has n-1 edges
Notice that C contains at least one edge t* that is not in T
why? because T is a tree, so acyclic
Notice that the weight of every such t* is at least the weight of
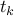
why? because t* does not form a cycle with edges
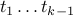 , which are all in T*, and yet Krusal picked
, which are all in T*, and yet Krusal picked
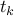 instead of t*
instead of t*
Notice that the weight of every such t* is at most the weight of
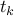
why? because T' = T* -t* +
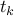 is a spanning tree, with weight
wt(T*) - wt(t*) + wt(
is a spanning tree, with weight
wt(T*) - wt(t*) + wt(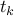 ), so if wt(t*) > wt(
), so if wt(t*) > wt(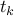 ) then
wt(T') < wt(T*), contradiction since T* is an MST
) then
wt(T') < wt(T*), contradiction since T* is an MST
so, for every such t*, wt(t*) = wt(
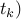
pick any t*, and let T' = T* -t* +
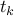
notice T' is an MST, with one more edge of T than T*
so repeating the above process as necessary, we eventually find an MST that contains all the edges of T, i.e. that equals T.
Kruskal mst completeness
recall: in each step of K's algm, any min-cost candidate edge can be added to the tree-so-far
call an mst kruskal if it is found by some execution of K's algm
claim: every mst is Kruskal
proof: by induction, contradiction form: consider an arbitrary mst T of a graph G. among all possible Kruskal executions on G, consider the one that yields the Kruskal mst K that maximizes the initial sequence of edges that are in T. say the sequence of edges of this execution is S=( k1, ..., kn-1) . so these edges form the mst K.
let q be smallest index such that kq is not in T.
T+kq has a unique cycle C. The edges of A={ k1, ..., kq } are picked by Kruskal, so acyclic, so C includes some edge not in A, say c.
kq is a max weight edge on C. But Kruskal picks kq before c, so kq weighs no more than c. So kq weighs the same as c.
So Kruskal could have picked c instead of kq. So some execution of Kruskal starts by selecting the edge sequence ( k1, ..., kq ) , contradicting our choice of K.
better UF
what is total cost of these operations? union(A B) union(B C) union(C D) ... union(Y Z) find(A) find(B) find(A) find(D) ... find(A) find(Z) motivation: keep UF trees shorter two options: union-by-rank, or find-grandparent-compress
def findGP(x, parents): # find, gp compress
px = parents[x]
if x==px: return x
gx = parents[px] # grandparent
while px != gx:
parents[x] = gx
x = px ; px = gx ; gx = parents[gx]
return px
def unionBR(v,w,P,R): # union by rank
rv,rw = myfind(v),myfind(w)
if R[rv] < R[rw]:
P[rv] = rw
elif R[rv] > R[rw]:
P[rw] = rv
else:
P[rv] = rw
R[rw] += 1
min cut of a graph: cut (S,V-S) that minimizes number of cross edges
brute force algorithm: consider all
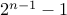 possible cuts
possible cutsKarger's randomized algm:
single iteration: Kruskal (except select edges at random), stop after
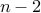 edges
edgeswith prob. at least
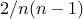 , cut is min
, cut is minperform
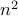 iterations, take smallest cut, with high prob is min
iterations, take smallest cut, with high prob is min
consider min cut, say size
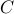
on first step,
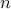 components
componentseach component incident
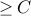 edges
edgesso
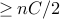 edges
edgesProb(picking edge from
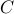 )
) 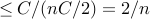
when
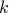 components
componentseach component incident
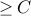 edges
edgesso
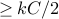 edges
edgesif so far have missed
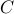 ,
,
Prob(picking edge from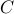 )
) 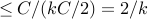
prob(avoiding
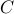 each time)
each time)
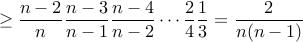
consider the graph with vertices A to F and edges AB AC BC DE DF EF plus AD
uniqe min cut {A B C} {D E F} has size 1 (edge AD)
probability that Karger's algorithm finds this cut?
to find this cut, must pick 2 edges from each of 2 triangles
prob that first edge from a triangle: 6/7
prob that second edge from a triangle: 5/6
two cases:
i) prob 2/5: one triangle has 1 edge remaining, the other 3
ii) prob 3/5: each triangle has 2 edges remaining
prob third edge from a triangle: i) 3/4 (one of 5 remaining edges would create a cycle if picked) ii) 4/5
prob fourth edge from a triangle: 2/3 (one of 4 remaining edges would create a cycle if picked)
total prob
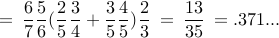
general bound ? 6-vertex graph, prob
import random
def myfind(x, parents):
if x==parents[x]: return x
return myfind(parents[x],parents)
def myunion(x,y,parents):
parents[x] = y
def nodeedgecount(G):
n,m = 0,0
for v in G:
n+= 1
for w in G[v]:
if v < w: m+= 1
return n,m
def edgelist(G):
L = []
for v in sorted(G):
for w in G[v]:
if v < w: L.append((v,w))
return L
def cutedges(E,P):
edgeset = []
for e in E:
if myfind(e[0],P) != myfind(e[1],P):
edgeset.append(e)
return edgeset
def makeparents(G):
p = []
for v in sorted(G):
p.append(v)
return p
def randomcut(G,n,E):
edgesadded = 0
P = makeparents(G)
for e in E:
x = myfind(e[0],P)
y = myfind(e[1],P)
if x != y:
myunion(x,y,P)
edgesadded += 1
if edgesadded == n-2:
return cutedges(E,P)
def showgoodcuts(G,t):
# iterate t times; show cut every time it improves
n,m = nodeedgecount(G)
E = edgelist(G)
bestcutsize = m+1 # initialize with impossible value
for j in range(t):
random.shuffle(E)
C = randomcut(G,n,E)
if len(C) < bestcutsize:
bestcut = C
bestcutsize = len(bestcut)
print 'iteration',j,bestcut
G = {0:[1,2,3], 1:[0,2,3], 2:[0,1,3,4], 3:[0,1,2,5],
4:[2,5,6,7], 5:[3,4,6,7], 6:[4,5,7], 7:[4,5,6] }
showgoodcuts(G,10)